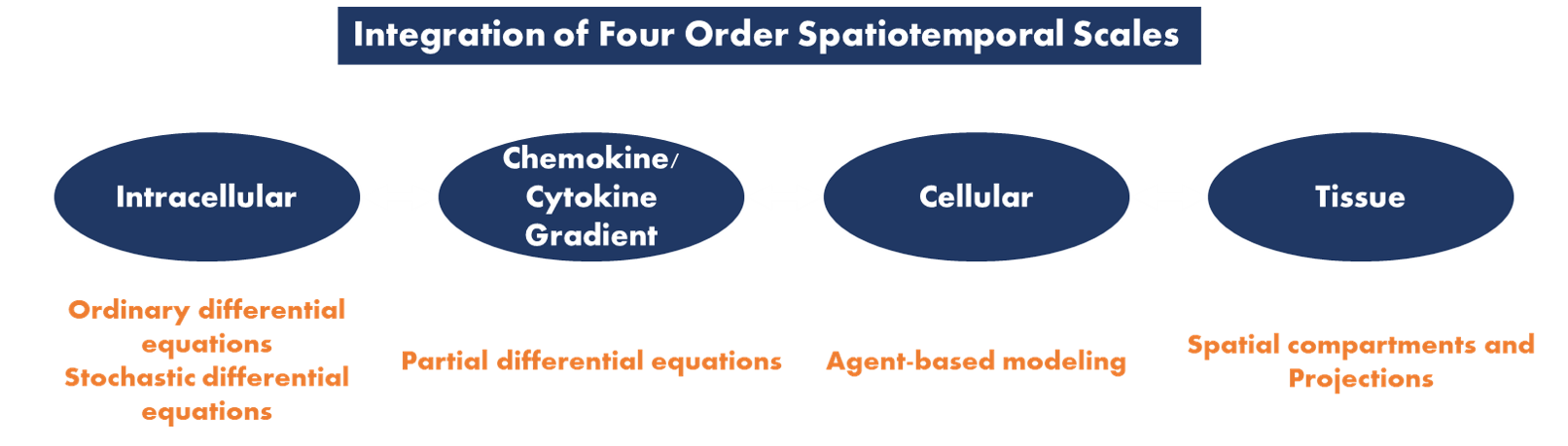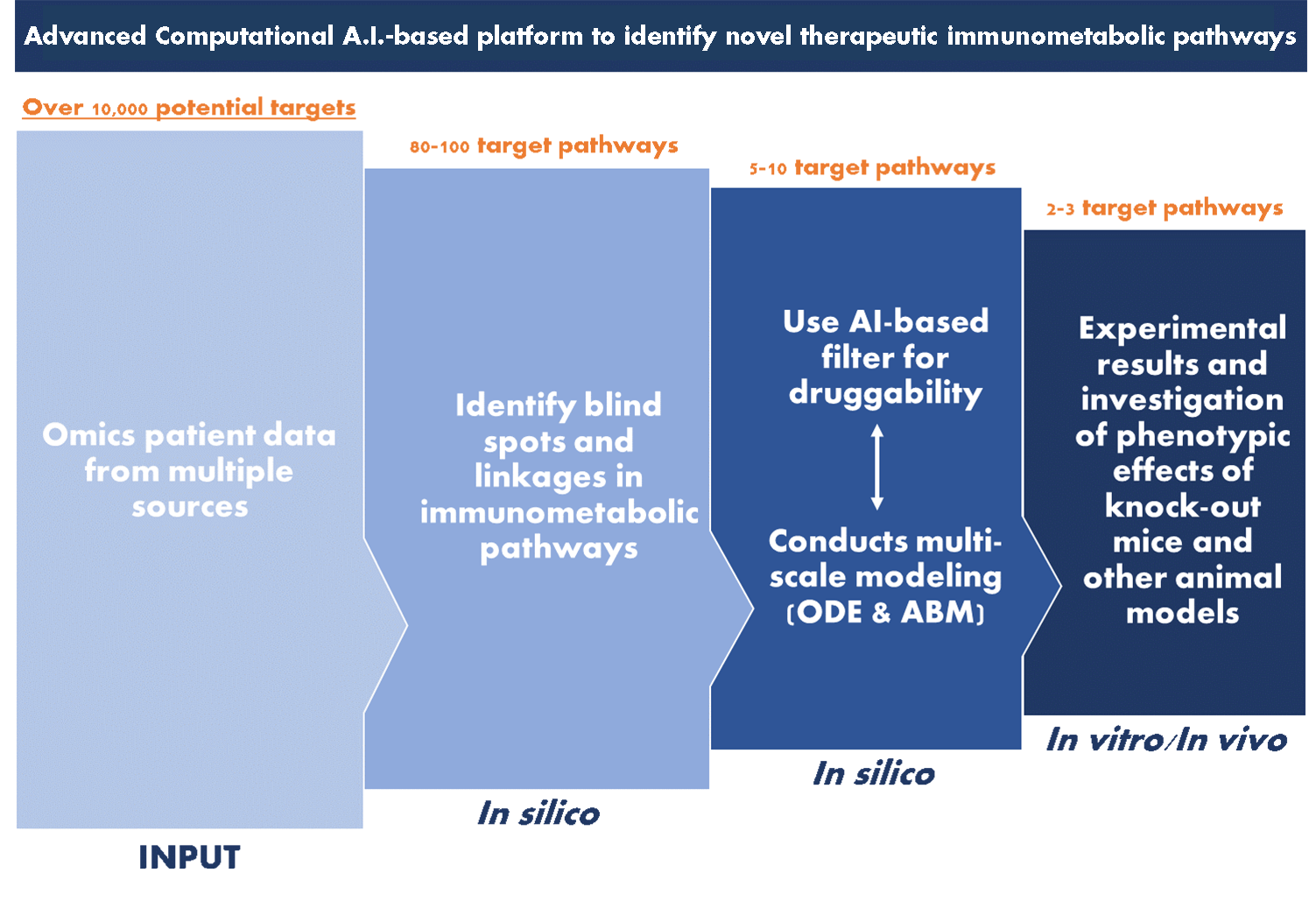
The NIMML Institute focuses on using AI techniques and computational modeling to further its position as a pioneer in the field of immunometabolism. A pipeline has been developed and utilized by NIMML to identify key connections between immunology and metabolism and target them effectively with potential drugs. By utilizing these techniques, NIMML stays on the cutting edge of immunometabolic research and drug development for immunoregulatory control and autoimmune therapy.
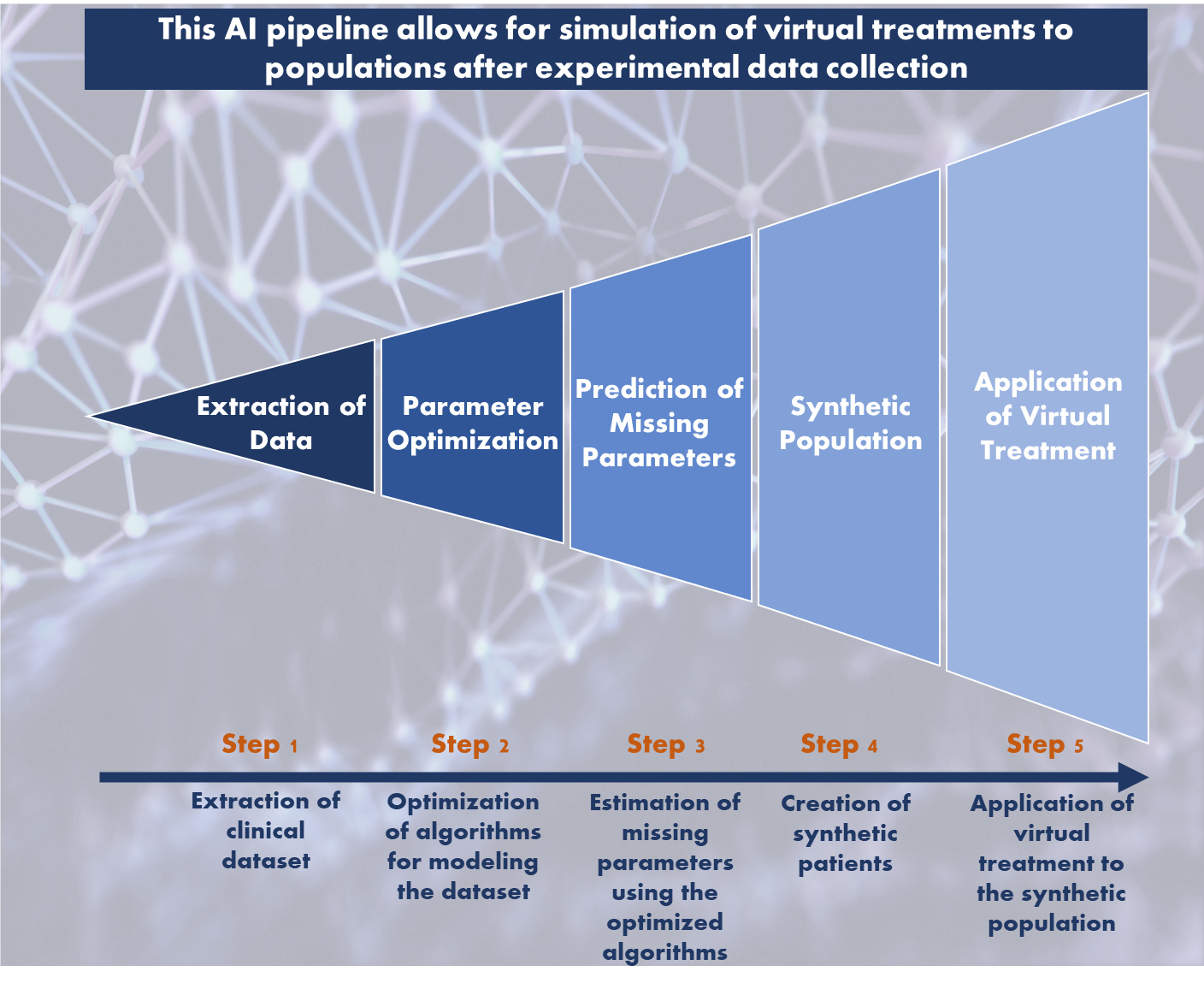
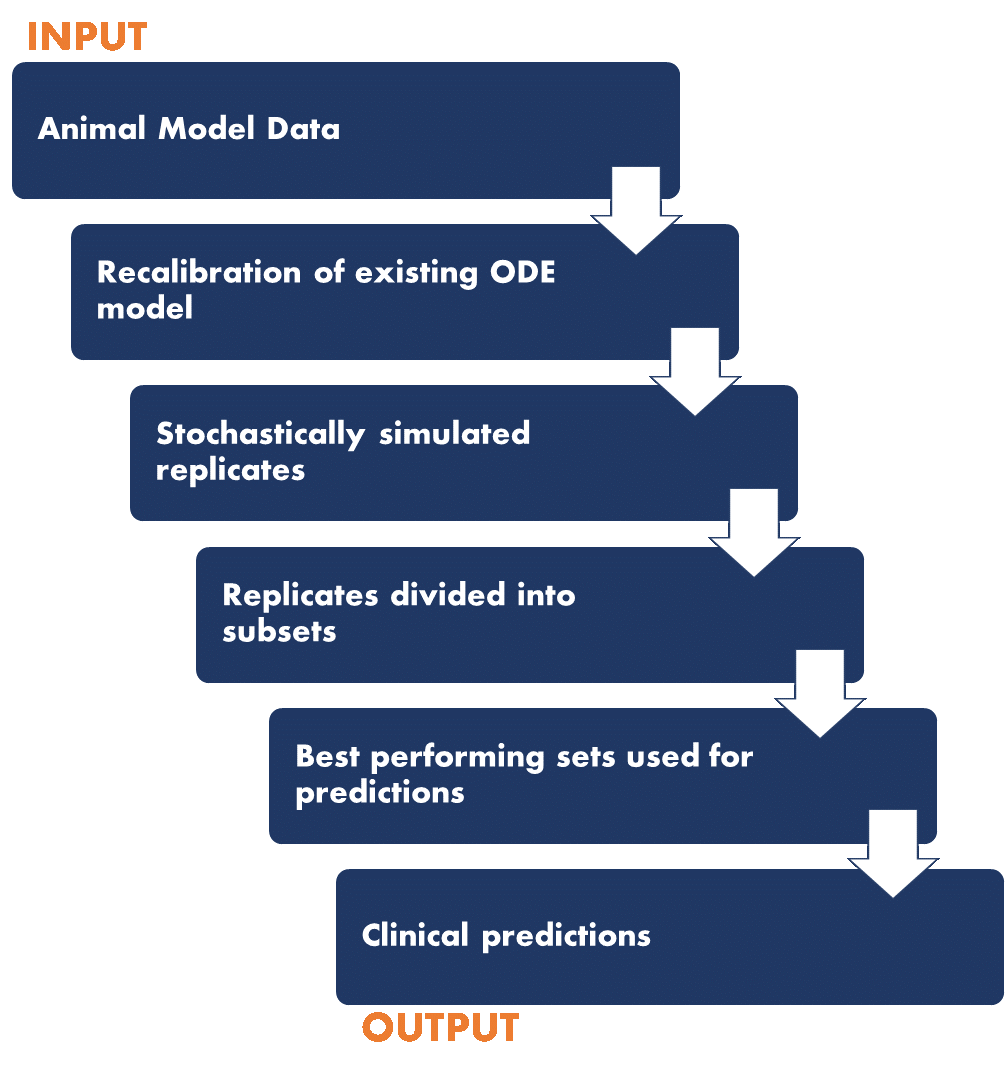
The NIMML Institute applied AI-based methods to develop a unique pipeline that produces useful insights for testing the clinical efficacy of IBD therapeutics in a randomized in silico trial. Crohn’s disease (CD) and ulcerative colitis (UC) are the two clinical manifestations of IBD, a widespread and debilitating autoimmune disease of unknown etiology afflicting 5 million people worlwide. Current therapies for IBD include the use of corticosteroids, antibiotics, immune-modulators, and anti-TNF-α biologics with limited efficacy and significant side effects. Thus, there is an unmet clinical need for safer and more effective oral therapeutics. To develop path to safer cures, we designed and implemented a novel integrated approach, based on the application advanced machine learning and AI algorithms. We created a large synthetic cohort of CD and UC patients and engineered these populations using immunological variables data derived from published clinical trials. For the proposed Phase III placebo-controlled, randomized in silico clinical trial, we simulated a test set of synthetic patients for five therapeutics. We randomly assigned five treatments -nutritional intervention (conjugated linoleic acid), a Phase II therapeutic (GED-0301 -Mongersen), a novel pre-clinical therapeutic targeting lanthionine synthetase C-like 2 (LANCL2), an anti-Tumor necrosis factor alpha (anti-TNF-α) antibody, and placebo to each synthetic patient. In CD trials, the change in Crohn’s disease activity index (CDAI) post 6 weeks was used to evaluate the effectiveness of current or investigational CD therapeutics. The change in CDAI score was based on the change in levels of biomarkers TNF-α (tumor necrosis factor-alpha) and IFN-γ (interferon gamma), which differed for every therapeutic intervention. For CD, the in silico study results demonstrated that therapeutics targeting LANCL2, ameliorated clinical disease in severe CD patients, with an average drop of 127 points from the initial CDAI score. The effectiveness of the LANCL2 therapeutics was comparable with anti-TNF-α antibodies. The analysis also highlighted that young patients (age < 40 years) respond significantly (P < 0.0001) better to treatment than older patients (age> 40 years).The study highlighted the value of in silico clinical trials for acceleration and improvement of drug development process. It sets a data-driven path for prediction of clinical outcome of investigational therapeutics forr autoimmune disease that leverages the advancements in machine learning and AI. The study opens up prospects that can aid in the future design of targeted precision medicine interventions combining biomarkers and therapeutic intervention for IBD patients with a long-term benefit of informed clinical development plans.