Advanced Computational Modeling of CD4+ T cell Immunometabolism
The Nutritional Immunology and Molecular Medicine Laboratory (NIMML) is pioneering the study of the immune responses as a massively and dynamically interacting system plus the development of advanced computational modeling approaches to solve the complex puzzle of immunity in infectious diseases. The NIMML has built the first advanced computational model of CD4+ T cell immunometabolism.
CD4+ T-cells, also known at T helper cells, orchestrate immune responses to pathogens by producing a mix of pleiotropic cytokines that will direct other cells of the adaptive and innate immune system on how to react to various stimuli.
Reprogramming CD4+ T cell responses through metabolic changes shows promise in developing novel host-based therapeutics for infectious diseases. Metabolic changes in CD4+ T cells are linked to immunological functions such as cell activation, cytokine production, and effector versus regulatory functionality during immune responses to infection and vaccination. Upon initiation of the immune response to infection, activated cells undergo significant metabolic changes to meet increased energy and structural requirements for cell proliferation. However, the immunometabolic mechanisms that control functional fates and disease outcomes during infection and vaccination remain largely unknown.
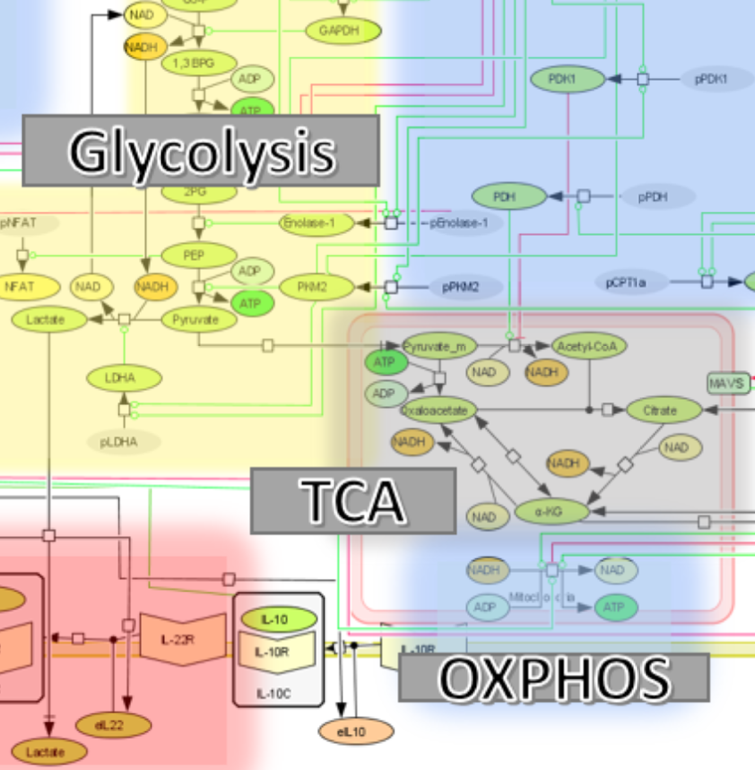
The immunological side of the model encompasses cytokines involved in CD4+ T cells differentiation and function, their corresponding receptors, canonical signaling pathways downstream of the cytokine receptors and transcriptional factors connected to key functional fates. The metabolic component of the model includes a wide range of metabolic processes including glycolysis, glutaminolysis, and fatty acid oxidation paying particular interest to the relationship between glycolysis, the citric acid cycle, and lactate production. By focusing on these pathways and how they interact with the immunological side of the biology, a comprehensive model has been developed, refined and validated. The model reflects the various subsets of effector and regulatory CD4+ T cells (naïve, Th1, Th17, and iTreg).
Analysis of the model focuses on the number of ATPs produced based on the cell type, and the ratio of ATPs produced via either pyruvate reduction to lactate (glycolytic pathway) or through oxidative phosphorylation. The model will be used to help identify and validate new immunometabolic therapeutic targets for infectious diseases.
Related press releases
Advanced Computational Modeling of the Gut for Biodefense
Immunometabolism – A Promising Gateway for Accelerated Drug Development
Applying Artificial Intelligence to Improve Drug Development
Artificial Intelligence: The Next Revolution in Healthcare and Precision Medicine
The NIMML Creates the First High-Resolution Agent-Based Model of the Gut
About NIMML
The NIMML Institute is a 501 (c) (3) non-profit public charity foundation focused on a transdisciplinary, team-science approach to precision medicine at the interface of immunology, inflammation, and metabolism. The NIMML Institute team has led numerous large-scale transdisciplinary projects and is dedicated to solving important societal problems by combining the expertise of immunologists, computational biologists, toxicologists, modelers, translational researchers, and molecular biologists. The Institute is headquartered in Blacksburg, VA. For more information, please visit www.nimml.org or contact pio@nimml.org.
